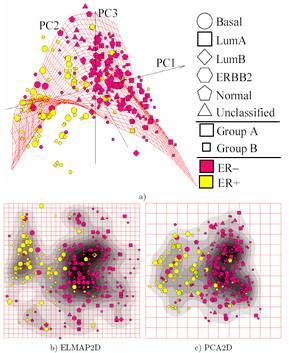
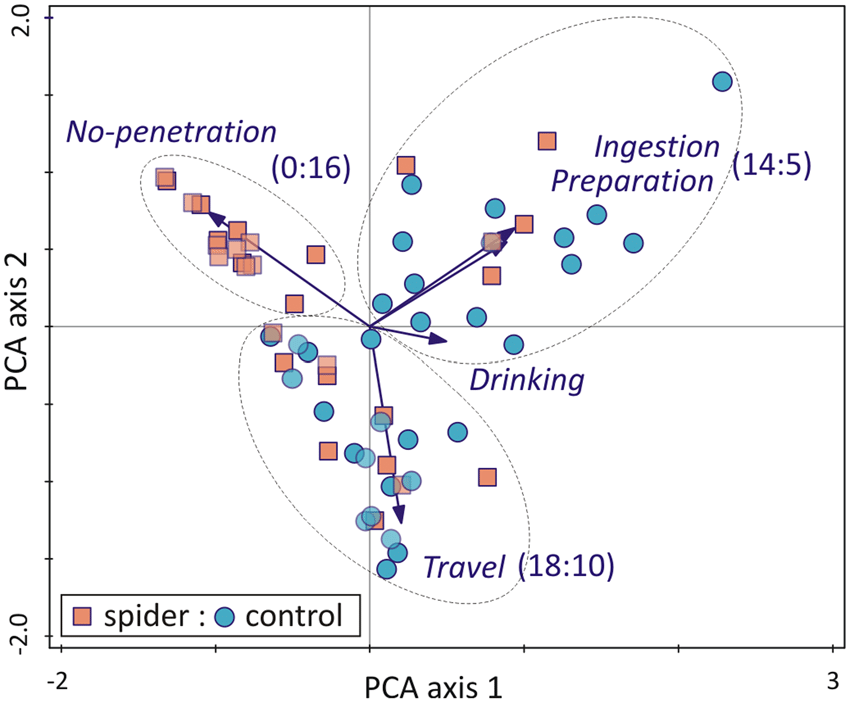
Quoted from: https://doi.org/10.1175%2F1520-0493%281987%29115%3C1825%3Aoaloma%3E2.0.co%3B2
PCA was invented in 1901 by Karl Pearson, as an analogue of the principal axis theorem in mechanics; it was later independently developed and named by Harold Hotelling in the 1930s. Depending on the field of application, it is also named the discrete Karhunen–Loève transform (KLT) in signal processing, the Hotelling transform in multivariate quality control, proper orthogonal decomposition (POD) in mechanical engineering, singular value decomposition (SVD) of X (Golub and Van Loan, 1983), eigenvalue decomposition (EVD) of XTX in linear algebra, factor analysis (for a discussion of the differences between PCA and factor analysis see Ch. 7 of Jolliffe's Principal Component Analysis), Eckart–Young theorem (Harman, 1960), or empirical orthogonal functions (EOF) in meteorological science, empirical eigenfunction decomposition (Sirovich, 1987), empirical component analysis (Lorenz, 1956), quasiharmonic modes (Brooks et al., 1988), spectral decomposition in noise and vibration, and empirical modal analysis in structural dynamics.
PCA can be thought of as fitting a p-dimensional ellipsoid to the data, where each axis of the ellipsoid represents a principal component. If some axis of the ellipsoid is small, then the variance along that axis is also small.
To find the axes of the ellipsoid, we must first subtract the mean of each variable from the dataset to center the data around the origin. Then, we compute the covariance matrix of the data and calculate the eigenvalues and corresponding eigenvectors of this covariance matrix. Then we must normalize each of the orthogonal eigenvectors to turn them into unit vectors. Once this is done, each of the mutually orthogonal, unit eigenvectors can be interpreted as an axis of the ellipsoid fitted to the data. This choice of basis will transform our covariance matrix into a diagonalised form with the diagonal elements representing the variance of each axis. The proportion of the variance that each eigenvector represents can be calculated by dividing the eigenvalue corresponding to that eigenvector by the sum of all eigenvalues.
PCA is defined as an orthogonal linear transformation that transforms the data to a new coordinate system such that the greatest variance by some scalar projection of the data comes to lie on the first coordinate (called the first principal component), the second greatest variance on the second coordinate, and so on.
Consider an {\displaystyle n\times p} data matrix, X, with column-wise zero empirical mean (the sample mean of each column has been shifted to zero), where each of the n rows represents a different repetition of the experiment, and each of the p columns gives a particular kind of feature (say, the results from a particular sensor).
data matrix, X, with column-wise zero empirical mean (the sample mean of each column has been shifted to zero), where each of the n rows represents a different repetition of the experiment, and each of the p columns gives a particular kind of feature (say, the results from a particular sensor).
Mathematically, the transformation is defined by a set of size {\displaystyle \ell } of p-dimensional vectors of weights or coefficients {\displaystyle \mathbf {w} _{(k)}=(w_{1},\dots ,w_{p})_{(k)}}
of p-dimensional vectors of weights or coefficients {\displaystyle \mathbf {w} _{(k)}=(w_{1},\dots ,w_{p})_{(k)}} that map each row vector {\displaystyle \mathbf {x} _{(i)}}
that map each row vector {\displaystyle \mathbf {x} _{(i)}} of X to a new vector of principal component scores {\displaystyle \mathbf {t} _{(i)}=(t_{1},\dots ,t_{l})_{(i)}}
of X to a new vector of principal component scores {\displaystyle \mathbf {t} _{(i)}=(t_{1},\dots ,t_{l})_{(i)}} , given by
, given by
- {\displaystyle {t_{k}}_{(i)}=\mathbf {x} _{(i)}\cdot \mathbf {w} _{(k)}\qquad \mathrm {for} \qquad i=1,\dots ,n\qquad k=1,\dots ,l}

in such a way that the individual variables {\displaystyle t_{1},\dots ,t_{\ell }} of t considered over the data set successively inherit the maximum possible variance from X, with each coefficient vector w constrained to be a unit vector (where {\displaystyle \ell }
of t considered over the data set successively inherit the maximum possible variance from X, with each coefficient vector w constrained to be a unit vector (where {\displaystyle \ell } is usually selected to be less than {\displaystyle p}
is usually selected to be less than {\displaystyle p} to reduce dimensionality).
to reduce dimensionality).
First component
In order to maximize variance, the first weight vector w(1) thus has to satisfy
- {\displaystyle \mathbf {w} _{(1)}={\underset {\Vert \mathbf {w} \Vert =1}{\operatorname {\arg \,max} }}\,\left\{\sum _{i}(t_{1})_{(i)}^{2}\right\}={\underset {\Vert \mathbf {w} \Vert =1}{\operatorname {\arg \,max} }}\,\left\{\sum _{i}\left(\mathbf {x} _{(i)}\cdot \mathbf {w} \right)^{2}\right\}}

Equivalently, writing this in matrix form gives
- {\displaystyle \mathbf {w} _{(1)}={\underset {\Vert \mathbf {w} \Vert =1}{\operatorname {\arg \,max} }}\,\{\Vert \mathbf {Xw} \Vert ^{2}\}={\underset {\Vert \mathbf {w} \Vert =1}{\operatorname {\arg \,max} }}\,\left\{\mathbf {w} ^{T}\mathbf {X^{T}} \mathbf {Xw} \right\}}

Since w(1) has been defined to be a unit vector, it equivalently also satisfies
- {\displaystyle \mathbf {w} _{(1)}={\operatorname {\arg \,max} }\,\left\{{\frac {\mathbf {w} ^{T}\mathbf {X^{T}} \mathbf {Xw} }{\mathbf {w} ^{T}\mathbf {w} }}\right\}}

The quantity to be maximised can be recognised as a Rayleigh quotient. A standard result for a positive semidefinite matrix such as XTX is that the quotient's maximum possible value is the largest eigenvalue of the matrix, which occurs when w is the corresponding eigenvector.
With w(1) found, the first principal component of a data vector x(i) can then be given as a score t1(i) = x(i) ⋅ w(1) in the transformed co-ordinates, or as the corresponding vector in the original variables, {x(i) ⋅ w(1)} w(1).
Further components
The kth component can be found by subtracting the first k − 1 principal components from X:
- {\displaystyle \mathbf {\hat {X}} _{k}=\mathbf {X} -\sum _{s=1}^{k-1}\mathbf {X} \mathbf {w} _{(s)}\mathbf {w} _{(s)}^{\rm {T}}}

and then finding the weight vector which extracts the maximum variance from this new data matrix
- {\displaystyle \mathbf {w} _{(k)}={\underset {\Vert \mathbf {w} \Vert =1}{\operatorname {arg\,max} }}\left\{\Vert \mathbf {\hat {X}} _{k}\mathbf {w} \Vert ^{2}\right\}={\operatorname {\arg \,max} }\,\left\{{\tfrac {\mathbf {w} ^{T}\mathbf {\hat {X}} _{k}^{T}\mathbf {\hat {X}} _{k}\mathbf {w} }{\mathbf {w} ^{T}\mathbf {w} }}\right\}}

It turns out that this gives the remaining eigenvectors of XTX, with the maximum values for the quantity in brackets given by their corresponding eigenvalues. Thus the weight vectors are eigenvectors of XTX.
The kth principal component of a data vector x(i) can therefore be given as a score tk(i) = x(i) ⋅ w(k) in the transformed co-ordinates, or as the corresponding vector in the space of the original variables, {x(i) ⋅ w(k)} w(k), where w(k) is the kth eigenvector of XTX.
The full principal components decomposition of X can therefore be given as
- {\displaystyle \mathbf {T} =\mathbf {X} \mathbf {W} }

where W is a p-by-p matrix of weights whose columns are the eigenvectors of XTX. The transpose of W is sometimes called the whitening or sphering transformation. Columns of W multiplied by the square root of corresponding eigenvalues, that is, eigenvectors scaled up by the variances, are called loadings in PCA or in Factor analysis.
Covariances
XTX itself can be recognised as proportional to the empirical sample covariance matrix of the dataset XT.
The sample covariance Q between two of the different principal components over the dataset is given by:
- {\displaystyle {\begin{aligned}Q(\mathrm {PC} _{(j)},\mathrm {PC} _{(k)})&\propto (\mathbf {X} \mathbf {w} _{(j)})^{T}(\mathbf {X} \mathbf {w} _{(k)})\\&=\mathbf {w} _{(j)}^{T}\mathbf {X} ^{T}\mathbf {X} \mathbf {w} _{(k)}\\&=\mathbf {w} _{(j)}^{T}\lambda _{(k)}\mathbf {w} _{(k)}\\&=\lambda _{(k)}\mathbf {w} _{(j)}^{T}\mathbf {w} _{(k)}\end{aligned}}}

where the eigenvalue property of w(k) has been used to move from line 2 to line 3. However eigenvectors w(j) and w(k) corresponding to eigenvalues of a symmetric matrix are orthogonal (if the eigenvalues are different), or can be orthogonalised (if the vectors happen to share an equal repeated value). The product in the final line is therefore zero; there is no sample covariance between different principal components over the dataset.
Another way to characterise the principal components transformation is therefore as the transformation to coordinates which diagonalise the empirical sample covariance matrix.
In matrix form, the empirical covariance matrix for the original variables can be written
- {\displaystyle \mathbf {Q} \propto \mathbf {X} ^{T}\mathbf {X} =\mathbf {W} \mathbf {\Lambda } \mathbf {W} ^{T}}

The empirical covariance matrix between the principal components becomes
- {\displaystyle \mathbf {W} ^{T}\mathbf {Q} \mathbf {W} \propto \mathbf {W} ^{T}\mathbf {W} \,\mathbf {\Lambda } \,\mathbf {W} ^{T}\mathbf {W} =\mathbf {\Lambda } }

where Λ is the diagonal matrix of eigenvalues λ(k) of XTX. λ(k) is equal to the sum of the squares over the dataset associated with each component k, that is, λ(k) = Σi tk2(i) = Σi (x(i) ⋅ w(k))2.